There are multiple possibilities for failed reactions.
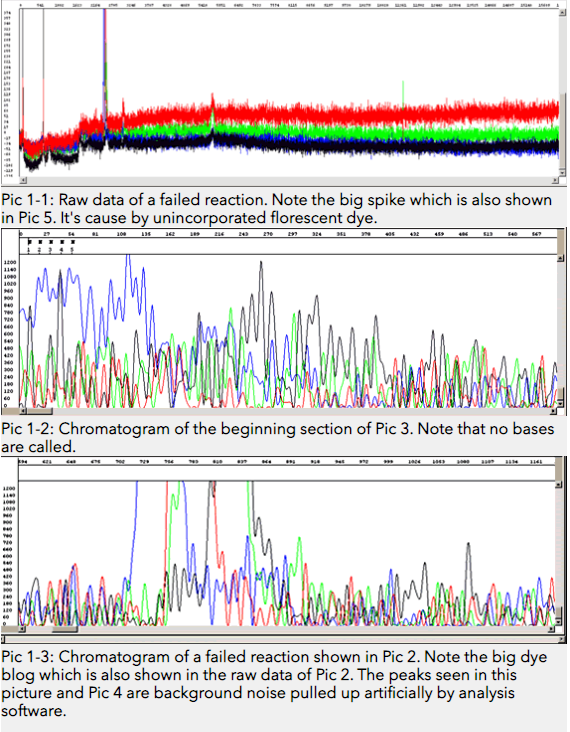
Possible Cause 1: Priming site not present
Solution: It most frequently happens with vector primers. If you've chosen on of our vector primers, make sure it is present in your vector. Double-check your plasmid maps/sequences.
If you've designed your own custom primer from previous sequence data, make sure you were using a reliable area of sequence - look for sharp, well-defined peaks with no ambiguity. Avoid areas where the peaks are broader and not well separated - this will occur towards the end of the sequence where the fragments are larger and the polymer cannot adequately resolve single nucleotides, causing inaccurate basecalling.
Possible Cause 2: Not enough or no DNA/Primer added
Solution: Double check your quantitations, stock concentrations, and dilutions. Check our FAQs for the amount of DNA we need. While our sequencers are very sensitive and can detect a range of DNA concentrations, there is still a "threshold" amount that must be reached to obtain any sequence data
Occasionally, either the primer or template is accidentally left out of a reaction. We try to immediately identify this type of mistake and repeat such reactions.
Possible Cause 3: Inhibitory contaminant
Solution: The cycle sequencing reaction used to amplify samples for automated sequencing is very sensitive to the presence of certain contaminants, some of which will completely inhibit our sequencing enzyme. Salts, EDTA, alcohol, protein, RNA, detergents, cesium and phenol are some of the most commonly seen contaminants that have a negative on sequencing enzyme. You may need to reprep your sample to sufficiently remove one or more inhibitory components to obtain any sequence data.
